Computational Biochemistry of Membrane Receptors
Computational simulation of biomolecules includes three dimensional modeling of receptors, simulation of their mechanisms of activation and elucidation of the molecular basis of their regulation by other molecules. Computational modeling has established as basic component to understand biochemistry, molecular pharmacology and designing drug-like compounds.
In our lab, we have specialised in developing computational methods to answer each of this points and apply our protocols to understand the signalling mechanism of membrane receptors, with a special focus on G protein-coupled receptors (GPCRs), arguably the largest family of drug targets. Our areas of interest include i) understanding and improving regulatory mechanisms by allosteric modulation, ii) design of novel chemical space with improved pharmacological profiles, iii) development of highly efficient physics-based modeling protocols, both to screen chemical compounds and to predicting the effect of pathogenic mutations.
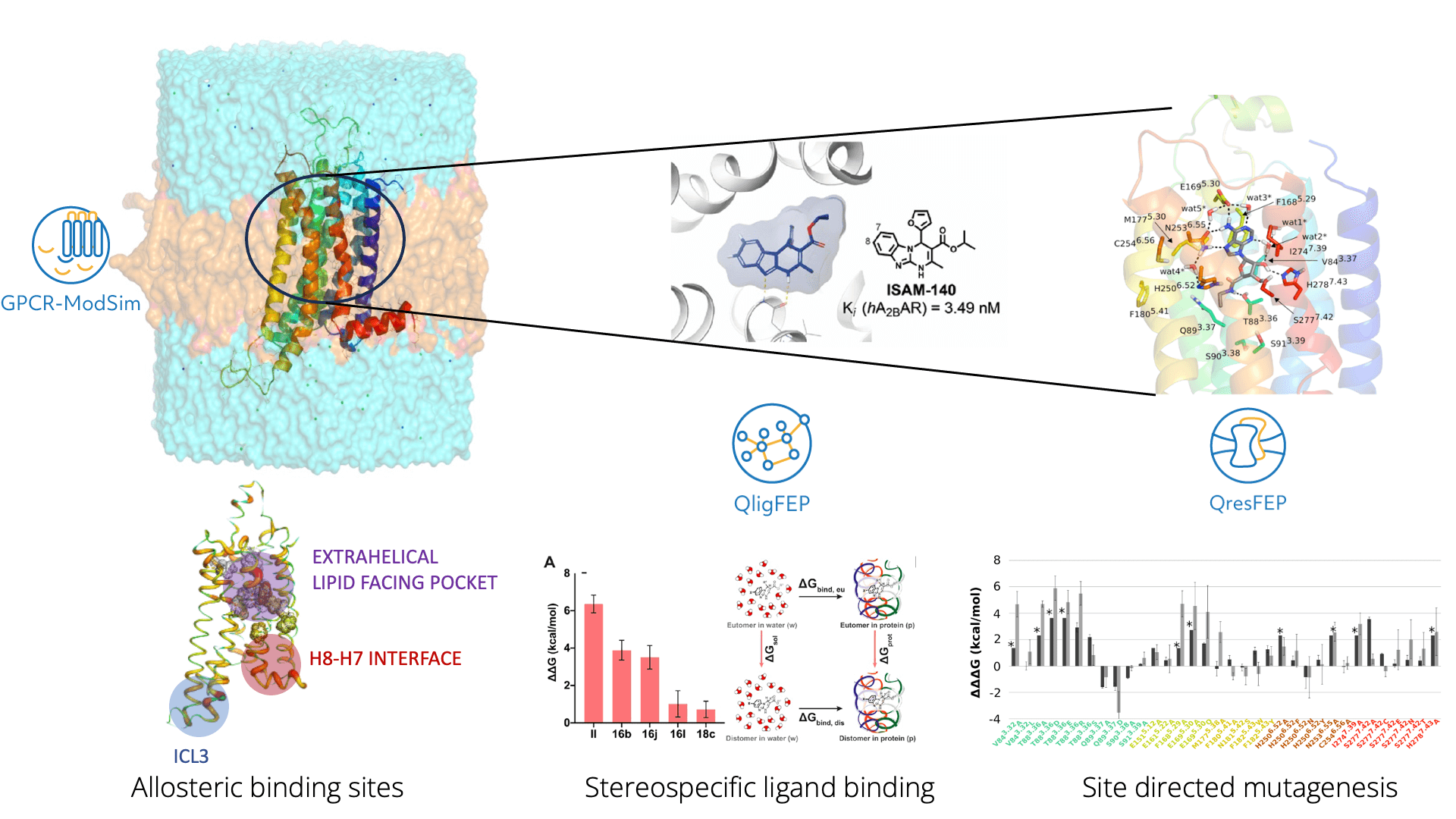
Our multiscale simulation methods allow us to locate allosteric binding sites (left, adenosine A1 receptor), determine the stereospecific binding mode of a series of adenosine A2B receptor antagonists, as immunological modulators in potential cancer treatments (center) or determine the effect of point mutations on ligand binding in the A2A receptor of the same family (right), a method with potential applications in predicting selectivity or resistances.

Laboratory Head
Email: h.g.teran@cinn.es
